Organic Molecule
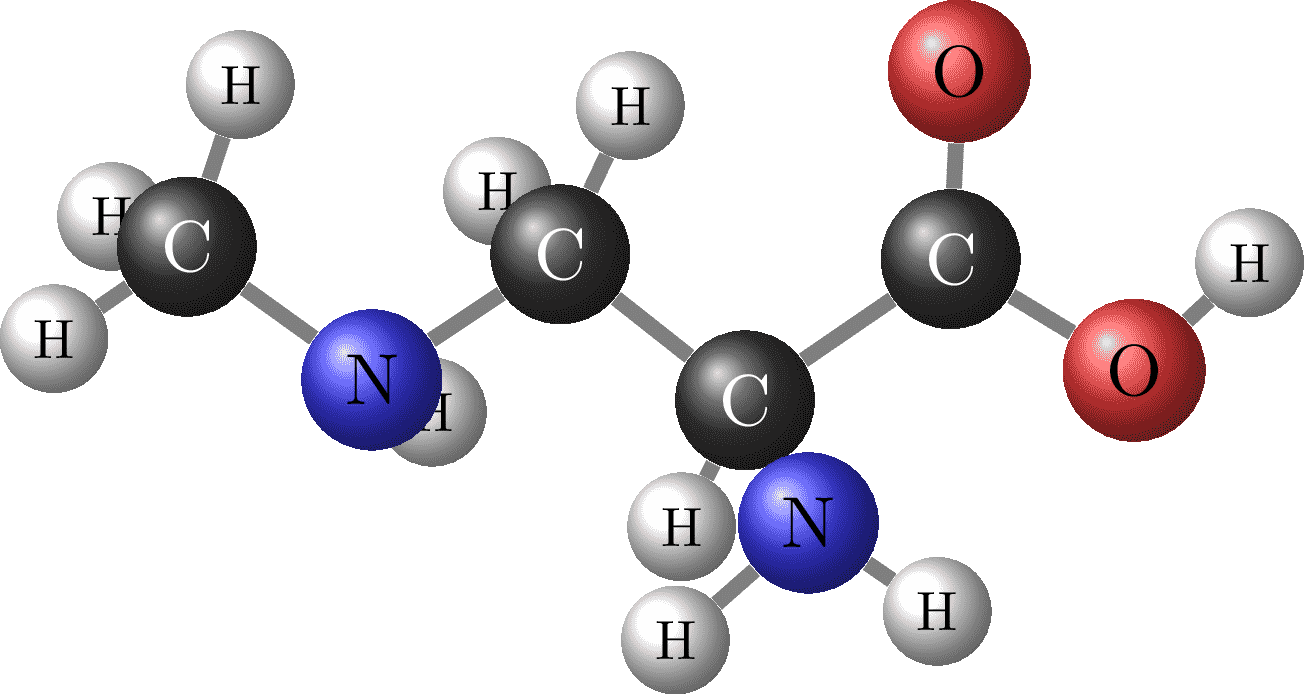
Beta-methylamino-DL-alanine - C4H10N2O2. Coordinates found on https://tex.stackexchange.com/q/453740.
Download
Code
organic-molecule.typ (213 lines)
organic-molecule.tex (33 lines)
Beta-methylamino-DL-alanine - C4H10N2O2. Coordinates found on https://tex.stackexchange.com/q/453740.
