Materials Informatics
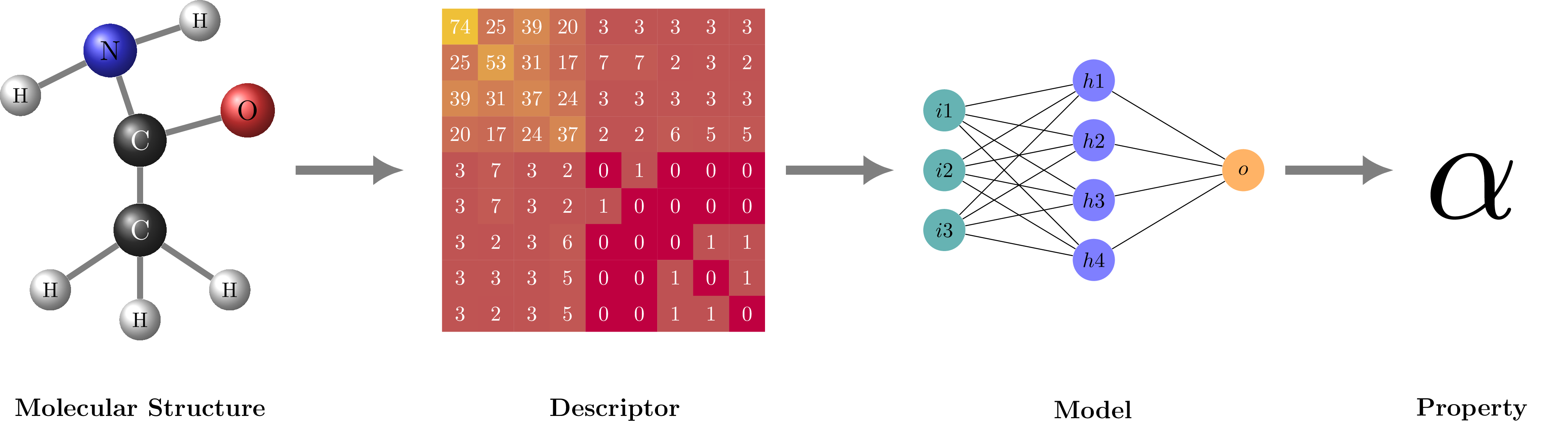
Structure-based materials informatics workflow. Inspired by fig. 1 in https://doi.org/10.1016/j.cpc.2019.106949.
Download
Code
materials-informatics.typ (342 lines)
materials-informatics.tex (81 lines)
Structure-based materials informatics workflow. Inspired by fig. 1 in https://doi.org/10.1016/j.cpc.2019.106949.
