Variational Autoencoder
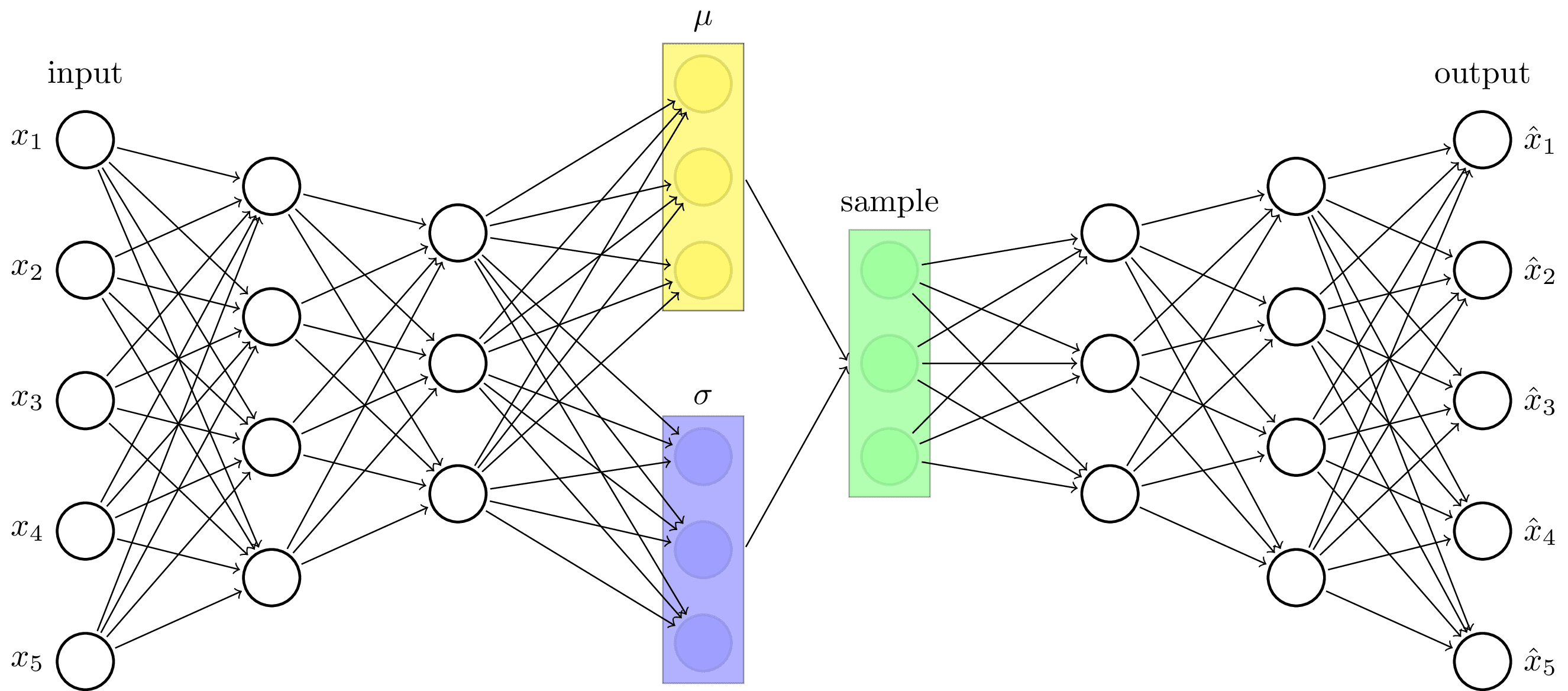
Variational autoencoder architecture. The earliest type of generative machine learning model. Inspired by https://towardsdatascience.com/intuitively-understanding-variational-autoencoders-1bfe67eb5daf.
Download
Code
variational-autoencoder.typ (143 lines)
variational-autoencoder.tex (78 lines)